Explaining and Predicting Two-Photon Absorption with Machine Learning
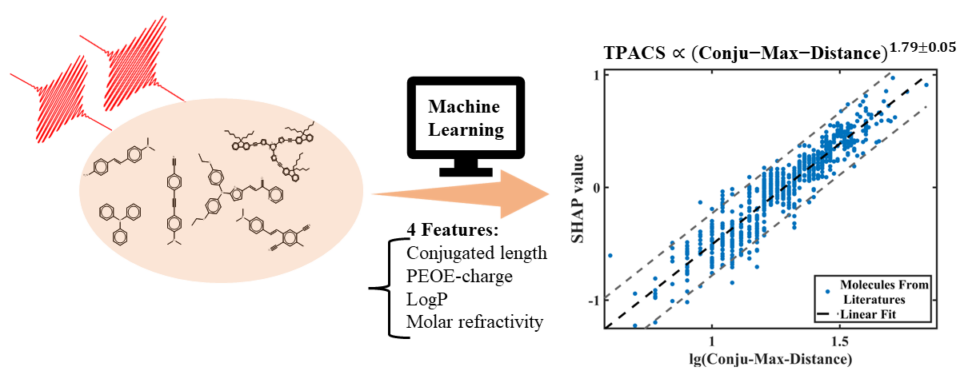
Materials can simultaneously absorb not just one but two photons and molecules with strong two-photon absorption (TPA) are important in many fields such as unconverted laser, photodynamic therapy, and 3D printing. In our work published in Advanced Science (open access), we use machine learning (ML) to quantitatively give insight into important factors affecting TPA which can be used in the design of such molecules. Our ML model can also fast and accurately predict the TPA of a molecule from its SMILES and is available in an open-source MLatom package which can be used for free on the XACS cloud computing service.
What’s TPA?
Two-photon absorption (TPA) is a special excitation process that involves two photons simultaneously, which leads to a non-linear correlation between the light intensity and its occurrence. As a result of its unusual characteristics, the TPA can be used in applications in many fields, e.g. upconverted laser, bio-imaging, photodynamic therapy, 3D printing, etc.
However, the design of new TPA molecules is not that easy: empirical conclusions are limited to some specific systems. While for quantum chemical methods, the computational cost and the accuracy are hard to balance.
Data set with experimental two-photon absorption measurements

Figure adopted from the manuscript.
In order to reliably establish key properties that influence molecular TPAs, we need as large data set as possible. Unfortunately, such data sets were not available in literature. Thus, one of the main contributions of our work is that we collected an experimental data set of 929 organic molecules from 275 literature reports. The data set contains the TPACSs, the SMILES, names of the molecules, wavelengths of the TPA test, TPA measurement methods, solvents, and DOI number of the source publication. In our work, a subset with 856 molecules with excitation wavelengths between 600-1100 nm is used in model training.
The interpretable ML-TPA model
We trained models for predicting TPA cross-section by developing an interpretable machine-learning method and our final model has at least comparable accuracy to DFT methods.
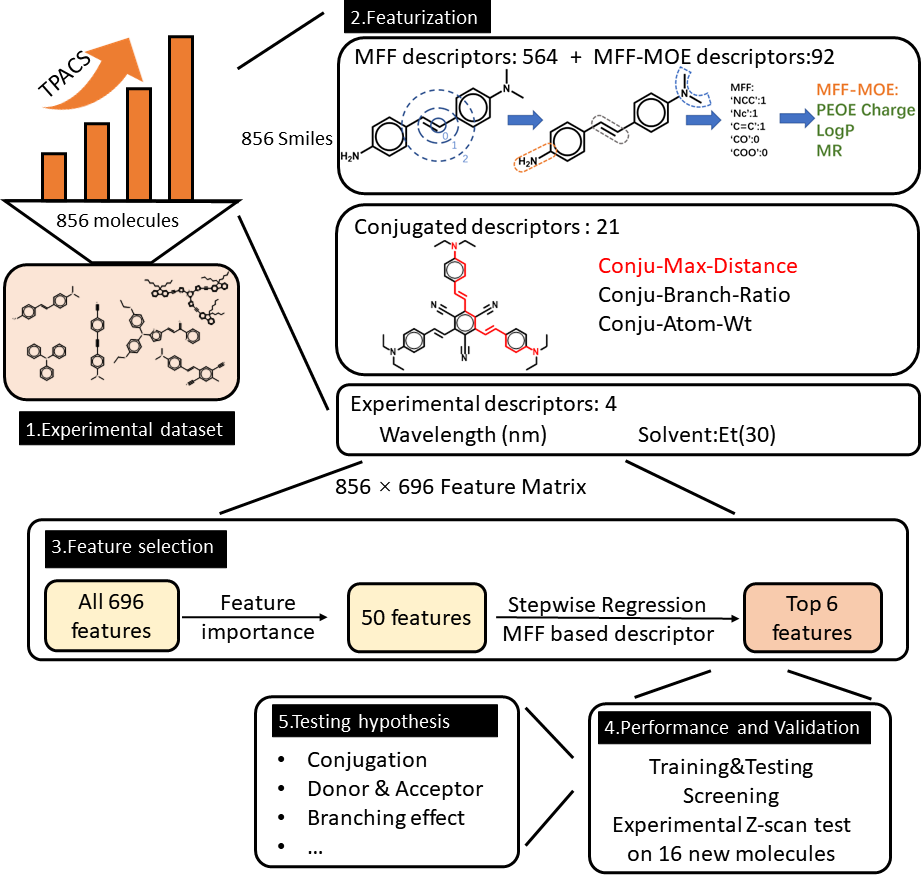
Figure adopted from the manuscript.
The major challenge of our work was feature selection – finding the most important descriptors. After thorough screening of features, we identified that many plausibly good features such as quantum mechanical descriptors (HOMO-LUMO gap, oscillator strengths…) were less important than several, relatively simple, key features. And those key features are:
- maximum conjugated length (Conju-Max-Distance. By far, the most important molecular feature)
- wavelength
- solvent characterized by ET(30) value
- maximum charge among charges of molecular fragments, defined by descriptor PEOE-Charge-Max (maximum PEOE charge)
- minimum logP value (LogP-Min)
- maximum molar refractivity (MR-Max)

Figure adopted from the manuscript.
With only these 6 features, the trained model can be basically as good as with 696 features:
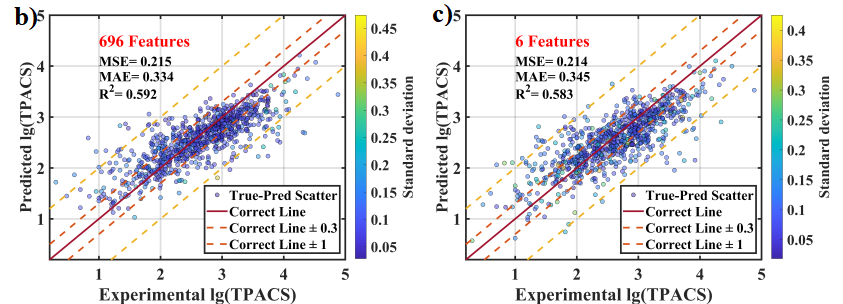
Figure adopted from the manuscript.
To further interpret our model, we performed the analysis on the SHAP values of different features. The SHAP value linearly indicates the contribution of a feature in the ML prediction, with value 0 meaning that the feature will not change the prediction from the average.
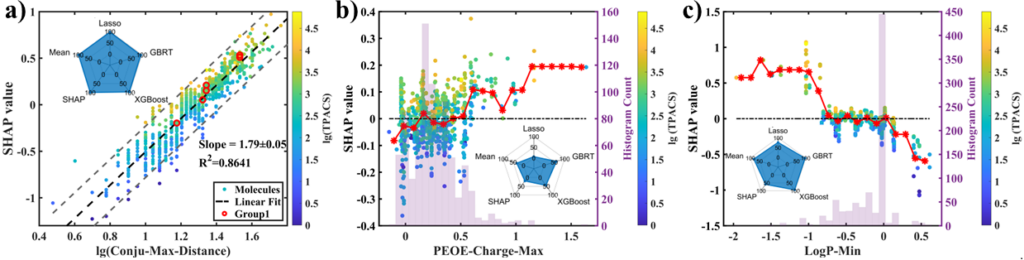
Figure adopted from the manuscript.
From the result of SHAP analysis, we’ve extracted many interesting insights. One of them is for the maximum conjugated length (Conju-Max-Distance, see the left panel in the above figure): the linear correlation between its log value and SHAP value was observed. Since the prediction of TPA cross-section is also in logarithmic scale, the following power law was inferred from the slope of 1.79±0.05:
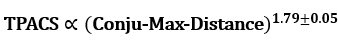
The donor-acceptor structure is also found to be important in TPA from the result of LogP-Min and PEOE-Charge-Max (the right and the middle panel in the figure above). The feature LogP-Min can mark the existence of highly polar groups, which are usually good electron donors/acceptors, and the lower the value is, the higher polarity it indicates. Its SHAP plot shows a negative correlation, which is consistent with the push-pull principle. The PEOE charge is obtained by summing up the Gasteiger charges of a molecular fragment, and the feature PEOE-Charge-Max supplements the description by identifying a positively charged conjugated carbon backbone that is connected to a strong electron-withdrawing group, as shown by the SHAP plot that adds a correction to the positively valued region.
Although many established concepts of the TPA structure-property relationship are validated by our results, multipolar DAn or ADn structure from the branching of the conjugated system was surprisingly found not beneficial for TPA from our analysis, which revises some of the previous beliefs.
How can I use ML-TPA?
Simple.
You can perform ML-TPA prediction by just providing the SMILES of your molecule with our MLatom package available on XACS cloud computing, please refer to the manual of ML-TPA.
Reference
- Yuming Su, Yiheng Dai, Yifan Zeng, Caiyun Wei, Yangtao Chen, Fuchun Ge, Peikun Zheng, Da Zhou*, Pavlo O. Dral*, Cheng Wang*. Interpretable Machine Learning of Two-Photon Absorption. Adv. Sci. 2023, 2204902. DOI: 10.1002/advs.202204902.
0 Comments on “Explaining and Predicting Two-Photon Absorption with Machine Learning”